Calculating Basic Population Genetic Statistics from SNP Data
Introduction
In this vignette, you will calculate basic population genetic statistics from SNP data using R packages. These statistics serve as exploratory analysis and require to work at the population level. We will calculate:
- Genetic diversity,
- Test Hardy Weinberg
- \(F_{is}\) and global \(F_{st}\).
The dataset used for these analyses are for lodgepole pine (Pinus contorta, Pinaceae), a plant species. More information on the dataset and the species is available from A. Eckert’s website. Here we use the dataset with no presupposed idea of interpreting the results in a biological way. To make the calculations faster, we work only with a subset of the full dataset.
Load necessary resources and packages
Workflow for SNP data
Import data
The data are stored in a text file (genotype=AA..). We will import the dataset into R as a data frame, and then convert the SNP data file into a “genind” object.
The dataset “Master_Pinus_data_genotype.txt” can be downloaded here.
The text file is a matrix of (550 rows x 3086 columns). It contains 4 extra columns: first column is the label of the individuals, the three other are description of the region, all the other columns are for the genotypes as (AA or AT…).
When you import the data into R, the data file needs to be in your working directory, or adjust the path in the read.table() invocation below accordingly.
Mydata <- read.table("Master_Pinus_data_genotype.txt", header = TRUE)
dim(Mydata) # Matrix of dimension 550x3086## [1] 550 3086To work with the data, we need to convert the R object returned by read.table() to a “genind” object. To achieve this, we create a matrix with only genotypes, and keep only a subset of the first 11 SNP loci (to make calculations faster). The result can then be converted to a “genind” object (for package adegent). The “genind” object can then easily be converted into a “hierfstat” (package hierfstat) object.
locus <- Mydata[, -c(1, 2, 3, 4, 17:3086)]
colnames(locus) <- gsub("\\.", "_", colnames(locus)) # locus names can't have "."
ind <- as.character(Mydata$tree_id) # labels of the individuals
population <- as.character(Mydata$state) # labels of the populations
Mydata1 <- df2genind(locus, ploidy = 2, ind.names = ind, pop = population, sep = "")
Mydata1## /// GENIND OBJECT /////////
##
## // 550 individuals; 12 loci; 24 alleles; size: 102.8 Kb
##
## // Basic content
## @tab: 550 x 24 matrix of allele counts
## @loc.n.all: number of alleles per locus (range: 2-2)
## @loc.fac: locus factor for the 24 columns of @tab
## @all.names: list of allele names for each locus
## @ploidy: ploidy of each individual (range: 2-2)
## @type: codom
## @call: df2genind(X = locus, sep = "", ind.names = ind, pop = population,
## ploidy = 2)
##
## // Optional content
## @pop: population of each individual (group size range: 4-177)## X0_10037_01_257 X0_10040_02_394 X0_10044_01_392 X0_10048_01_60
## 2 2 2 2
## X0_10051_02_166 X0_10054_01_402 X0_10067_03_111 X0_10079_02_168
## 2 2 2 2
## X0_10112_01_169 X0_10113_01_119 X0_10116_01_165 X0_10151_01_86
## 2 2 2 2Genetic diversity (observed and expected heterozygosity)
With adegenet:
##
## // Number of individuals: 550
## // Group sizes: 10 73 177 58 169 7 10 24 9 9 4
## // Number of alleles per locus: 2 2 2 2 2 2 2 2 2 2 2 2
## // Number of alleles per group: 22 24 24 23 24 20 19 23 21 19 18
## // Percentage of missing data: 1.62 %
## // Observed heterozygosity: 0.42 0.49 0.46 0.4 0.12 0.47 0.07 0.08 0.11 0.46 0.04 0.14
## // Expected heterozygosity: 0.41 0.5 0.5 0.45 0.13 0.5 0.07 0.08 0.11 0.44 0.04 0.16## [1] "n" "n.by.pop" "loc.n.all" "pop.n.all" "NA.perc" "Hobs"
## [7] "Hexp"plot(div$Hobs, xlab="Loci number", ylab="Observed Heterozygosity",
main="Observed heterozygosity per locus")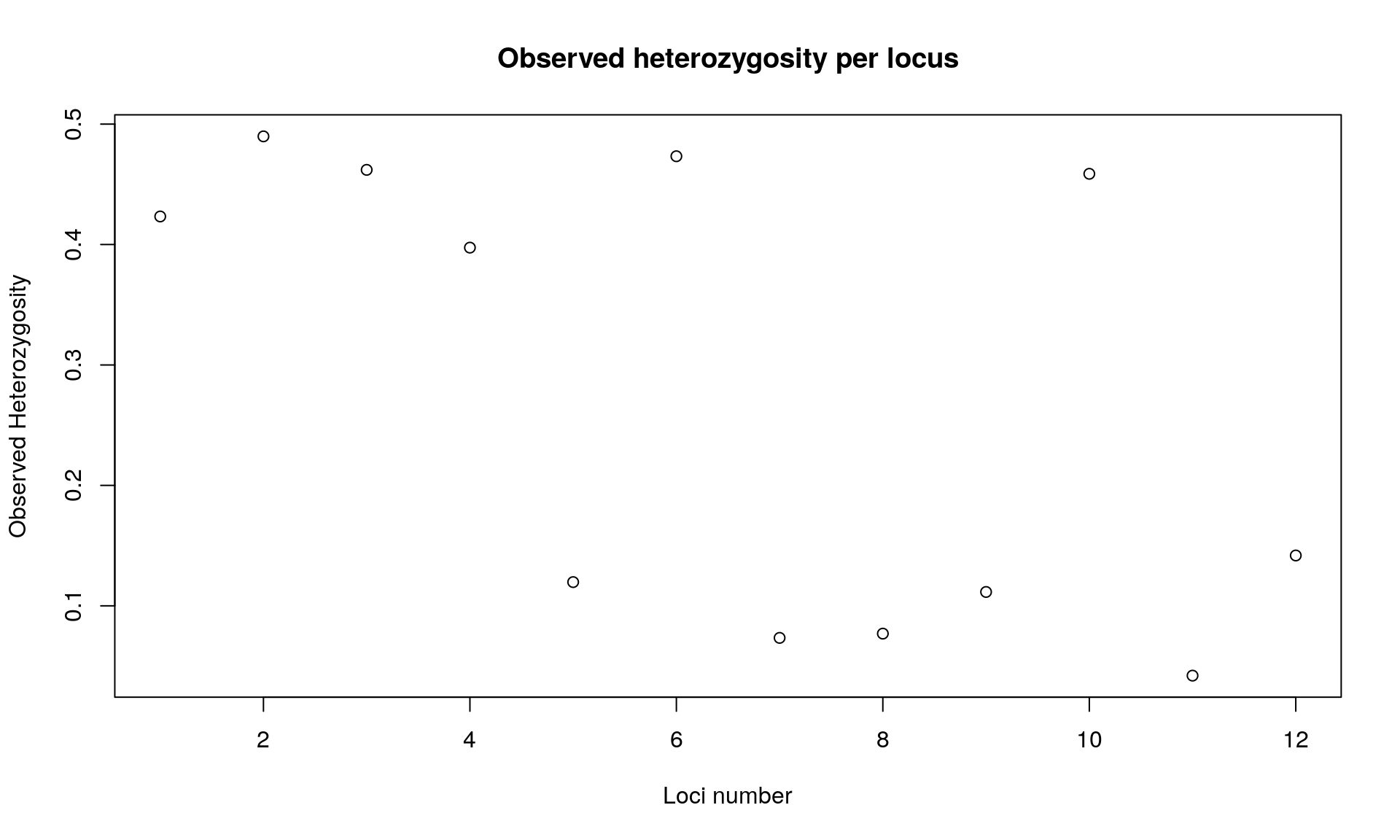
plot(div$Hobs,div$Hexp, xlab="Hobs", ylab="Hexp",
main="Expected heterozygosity as a function of observed heterozygosity per locus")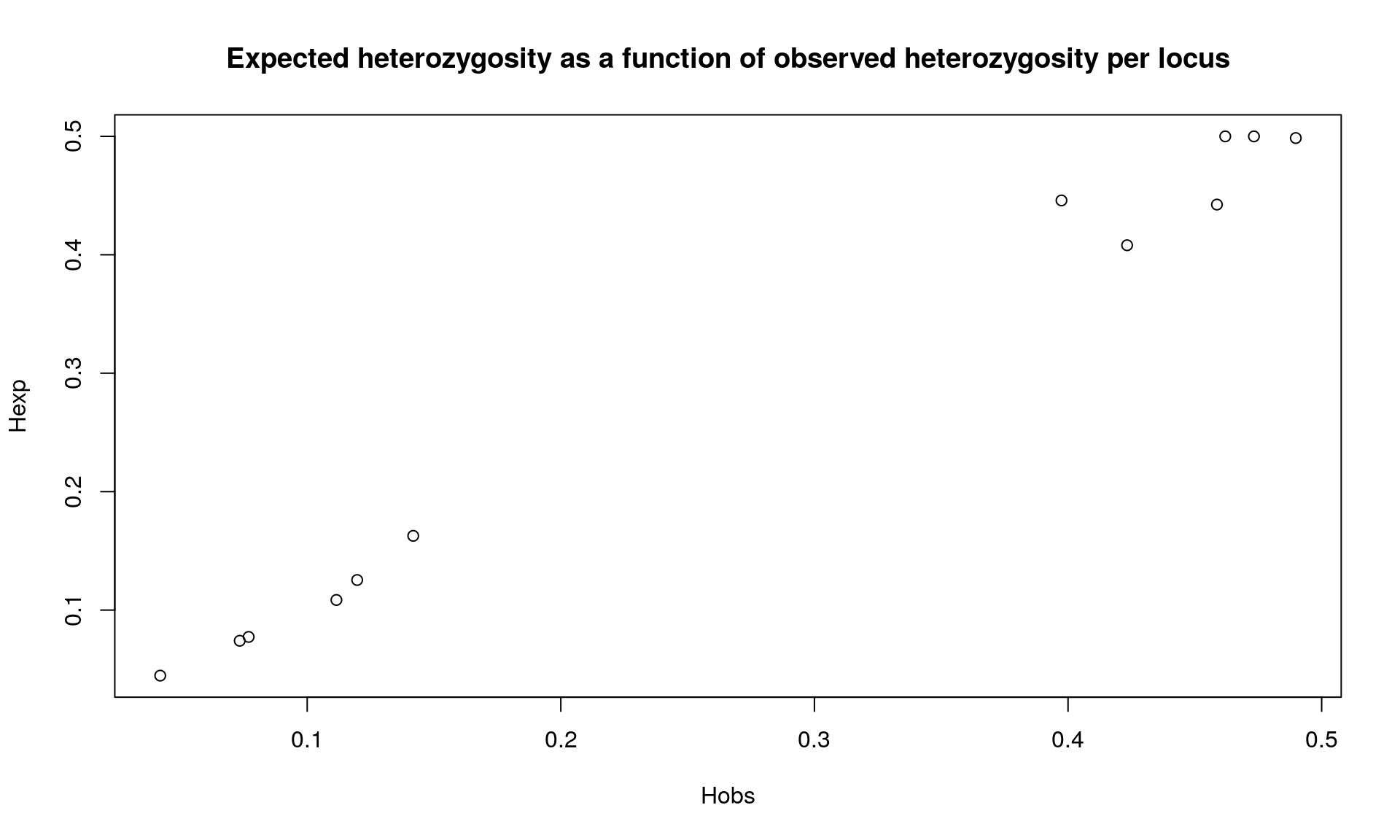
##
## Bartlett test of homogeneity of variances
##
## data: list(div$Hexp, div$Hobs)
## Bartlett's K-squared = 0.011457, df = 1, p-value = 0.9148We observed that heterozygosity varies among loci. We observed no difference between expected and observed heterozygosity.
Basic statistics with hierfstat. Populations are states. The function basic.stats() provides the observed heterozygosity (\(H_o\)), mean gene diversities within population (\(H_s\)), \(F_{is}\), and \(F_{st}\). The function boot.ppfis() provides confidence interval for \(F_{is}\). The function indpca() does an PCA on the centered matrix of individuals’ allele frequencies.
## [1] "n.ind.samp" "pop.freq" "Ho" "Hs" "Fis"
## [6] "perloc" "overall"## $call
## boot.ppfis(dat = Mydata2)
##
## $fis.ci
## ll hl
## 1 -0.0131 0.2944
## 2 -0.0515 0.0509
## 3 0.0202 0.0930
## 4 -0.0418 0.0829
## 5 -0.0255 0.0655
## 6 -0.5342 0.1645
## 7 -0.2003 0.1490
## 8 -0.2172 -0.0779
## 9 -0.1490 0.3288
## 10 0.0196 0.3683
## 11 -0.4238 -0.1538
Testing for Hardy-Weinberg Equilibrium
We used the function hw.test() from the pegas package.
## chi^2 df Pr(chi^2 >) Pr.exact
## X0_10037_01_257 0.75465490 1 0.385006451 0.450
## X0_10040_02_394 0.16628227 1 0.683437231 0.727
## X0_10044_01_392 2.96053195 1 0.085319867 0.094
## X0_10048_01_60 6.39076800 1 0.011471539 0.019
## X0_10051_02_166 1.12035389 1 0.289842265 0.283
## X0_10054_01_402 1.54582494 1 0.213752853 0.232
## X0_10067_03_111 0.04868128 1 0.825374035 0.562
## X0_10079_02_168 0.01579313 1 0.899992594 0.597
## X0_10112_01_169 0.41126025 1 0.521330544 1.000
## X0_10113_01_119 0.74248768 1 0.388865151 0.418
## X0_10116_01_165 1.87098306 1 0.171362531 0.242
## X0_10151_01_86 8.94232481 1 0.002786379 0.002We get for each locus a test of significance of the null hypothesis: \(H_0\) the locus is in HW equilibrium in the population. All but one locus are in HW equilibrium.
Conclusion
What did we learn today?
In this vignette, we learned how to explore the patterns of genetic diversity and how to estimate the level of genetic differentiation in one population. Also, you have an idea of potential violations of the dataset to the null Wright-Fischer model.
What is next?
You may now want to move into looking into population differentiation in more detail (See Calculating genetic differentiation and clustering methods from SNP data)
References
Eckert, A. J., A. D. Bower, S. C. González-Martínez, J. L. Wegrzyn, G. Coop and D. B. Neale. 2010. Back to nature: Ecological genomics of loblolly pine (Pinus taeda, Pinaceae). Molecular Ecology 19: 3789-3805. doi:[10.1111/j.1365-294X.2010.04698.x](http://dx.doi.org/10.1111/j.1365-294X.2010.04698.x)
Contributors
- Stéphanie Manel (Author)
- Zhian Kamvar (edits)
Session Information
This shows us useful information for reproducibility. Of particular importance are the versions of R and the packages used to create this workflow. It is considered good practice to record this information with every analysis.
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 3.6.1 (2019-07-05)
## os Debian GNU/Linux 9 (stretch)
## system x86_64, linux-gnu
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Etc/UTC
## date 2019-09-16
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────
## package * version date lib source
## ade4 * 1.7-13 2018-08-31 [1] CRAN (R 3.6.1)
## adegenet * 2.1.1 2018-02-02 [1] CRAN (R 3.6.1)
## ape * 5.3 2019-03-17 [1] CRAN (R 3.6.1)
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 3.6.1)
## backports 1.1.4 2019-04-10 [1] CRAN (R 3.6.1)
## boot 1.3-22 2019-04-02 [2] CRAN (R 3.6.1)
## callr 3.3.1 2019-07-18 [1] CRAN (R 3.6.1)
## class 7.3-15 2019-01-01 [2] CRAN (R 3.6.1)
## classInt 0.4-1 2019-08-06 [1] CRAN (R 3.6.1)
## cli 1.1.0 2019-03-19 [1] CRAN (R 3.6.1)
## cluster 2.1.0 2019-06-19 [2] CRAN (R 3.6.1)
## coda 0.19-3 2019-07-05 [1] CRAN (R 3.6.1)
## colorspace 1.4-1 2019-03-18 [1] CRAN (R 3.6.1)
## crayon 1.3.4 2017-09-16 [1] CRAN (R 3.6.1)
## DBI 1.0.0 2018-05-02 [1] CRAN (R 3.6.1)
## deldir 0.1-23 2019-07-31 [1] CRAN (R 3.6.1)
## desc 1.2.0 2018-05-01 [1] CRAN (R 3.6.1)
## devtools 2.2.0 2019-09-07 [1] CRAN (R 3.6.1)
## digest 0.6.20 2019-07-04 [1] CRAN (R 3.6.1)
## dplyr 0.8.3 2019-07-04 [1] CRAN (R 3.6.1)
## DT 0.8 2019-08-07 [1] CRAN (R 3.6.1)
## e1071 1.7-2 2019-06-05 [1] CRAN (R 3.6.1)
## ellipsis 0.2.0.1 2019-07-02 [1] CRAN (R 3.6.1)
## evaluate 0.14 2019-05-28 [1] CRAN (R 3.6.1)
## expm 0.999-4 2019-03-21 [1] CRAN (R 3.6.1)
## fs 1.3.1 2019-05-06 [1] CRAN (R 3.6.1)
## gdata 2.18.0 2017-06-06 [1] CRAN (R 3.6.1)
## ggplot2 3.2.1 2019-08-10 [1] CRAN (R 3.6.1)
## glue 1.3.1 2019-03-12 [1] CRAN (R 3.6.1)
## gmodels 2.18.1 2018-06-25 [1] CRAN (R 3.6.1)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 3.6.1)
## gtools 3.8.1 2018-06-26 [1] CRAN (R 3.6.1)
## hierfstat * 0.04-22 2015-12-04 [1] CRAN (R 3.6.1)
## htmltools 0.3.6 2017-04-28 [1] CRAN (R 3.6.1)
## htmlwidgets 1.3 2018-09-30 [1] CRAN (R 3.6.1)
## httpuv 1.5.2 2019-09-11 [1] CRAN (R 3.6.1)
## igraph 1.2.4.1 2019-04-22 [1] CRAN (R 3.6.1)
## KernSmooth 2.23-15 2015-06-29 [2] CRAN (R 3.6.1)
## knitr 1.24 2019-08-08 [1] CRAN (R 3.6.1)
## later 0.8.0 2019-02-11 [1] CRAN (R 3.6.1)
## lattice 0.20-38 2018-11-04 [2] CRAN (R 3.6.1)
## lazyeval 0.2.2 2019-03-15 [1] CRAN (R 3.6.1)
## LearnBayes 2.15.1 2018-03-18 [1] CRAN (R 3.6.1)
## magrittr 1.5 2014-11-22 [1] CRAN (R 3.6.1)
## MASS 7.3-51.4 2019-03-31 [2] CRAN (R 3.6.1)
## Matrix 1.2-17 2019-03-22 [2] CRAN (R 3.6.1)
## memoise 1.1.0 2017-04-21 [1] CRAN (R 3.6.1)
## mgcv 1.8-28 2019-03-21 [2] CRAN (R 3.6.1)
## mime 0.7 2019-06-11 [1] CRAN (R 3.6.1)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 3.6.1)
## nlme 3.1-140 2019-05-12 [2] CRAN (R 3.6.1)
## pegas * 0.11 2018-07-09 [1] CRAN (R 3.6.1)
## permute 0.9-5 2019-03-12 [1] CRAN (R 3.6.1)
## pillar 1.4.2 2019-06-29 [1] CRAN (R 3.6.1)
## pkgbuild 1.0.5 2019-08-26 [1] CRAN (R 3.6.1)
## pkgconfig 2.0.2 2018-08-16 [1] CRAN (R 3.6.1)
## pkgload 1.0.2 2018-10-29 [1] CRAN (R 3.6.1)
## plyr 1.8.4 2016-06-08 [1] CRAN (R 3.6.1)
## prettyunits 1.0.2 2015-07-13 [1] CRAN (R 3.6.1)
## processx 3.4.1 2019-07-18 [1] CRAN (R 3.6.1)
## promises 1.0.1 2018-04-13 [1] CRAN (R 3.6.1)
## ps 1.3.0 2018-12-21 [1] CRAN (R 3.6.1)
## purrr 0.3.2 2019-03-15 [1] CRAN (R 3.6.1)
## R6 2.4.0 2019-02-14 [1] CRAN (R 3.6.1)
## Rcpp 1.0.2 2019-07-25 [1] CRAN (R 3.6.1)
## remotes 2.1.0 2019-06-24 [1] CRAN (R 3.6.1)
## reshape2 1.4.3 2017-12-11 [1] CRAN (R 3.6.1)
## rlang 0.4.0 2019-06-25 [1] CRAN (R 3.6.1)
## rmarkdown 1.15 2019-08-21 [1] CRAN (R 3.6.1)
## rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.6.1)
## scales 1.0.0 2018-08-09 [1] CRAN (R 3.6.1)
## seqinr 3.6-1 2019-09-07 [1] CRAN (R 3.6.1)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 3.6.1)
## sf 0.7-7 2019-07-24 [1] CRAN (R 3.6.1)
## shiny 1.3.2 2019-04-22 [1] CRAN (R 3.6.1)
## sp 1.3-1 2018-06-05 [1] CRAN (R 3.6.1)
## spData 0.3.0 2019-01-07 [1] CRAN (R 3.6.1)
## spdep 1.1-2 2019-04-05 [1] CRAN (R 3.6.1)
## stringi 1.4.3 2019-03-12 [1] CRAN (R 3.6.1)
## stringr 1.4.0 2019-02-10 [1] CRAN (R 3.6.1)
## testthat 2.2.1 2019-07-25 [1] CRAN (R 3.6.1)
## tibble 2.1.3 2019-06-06 [1] CRAN (R 3.6.1)
## tidyselect 0.2.5 2018-10-11 [1] CRAN (R 3.6.1)
## units 0.6-4 2019-08-22 [1] CRAN (R 3.6.1)
## usethis 1.5.1 2019-07-04 [1] CRAN (R 3.6.1)
## vegan 2.5-6 2019-09-01 [1] CRAN (R 3.6.1)
## withr 2.1.2 2018-03-15 [1] CRAN (R 3.6.1)
## xfun 0.9 2019-08-21 [1] CRAN (R 3.6.1)
## xtable 1.8-4 2019-04-21 [1] CRAN (R 3.6.1)
## yaml 2.2.0 2018-07-25 [1] CRAN (R 3.6.1)
##
## [1] /usr/local/lib/R/site-library
## [2] /usr/local/lib/R/library